# Selecting columns
Fertspm = FertSp.iloc[:, [1, 9, 15, 16]] # Adjust for zero-based index
NoFertspm = NoFertSp.iloc[:, [1, 9, 15, 16]]
# Merging data
Combined = pd.merge(Fertspm, NoFertspm, on="NAME", how="outer").fillna(0)
# Remove specific species
Combined = Combined[Combined['NAME'] != "Streptomyces griseofuscus"]
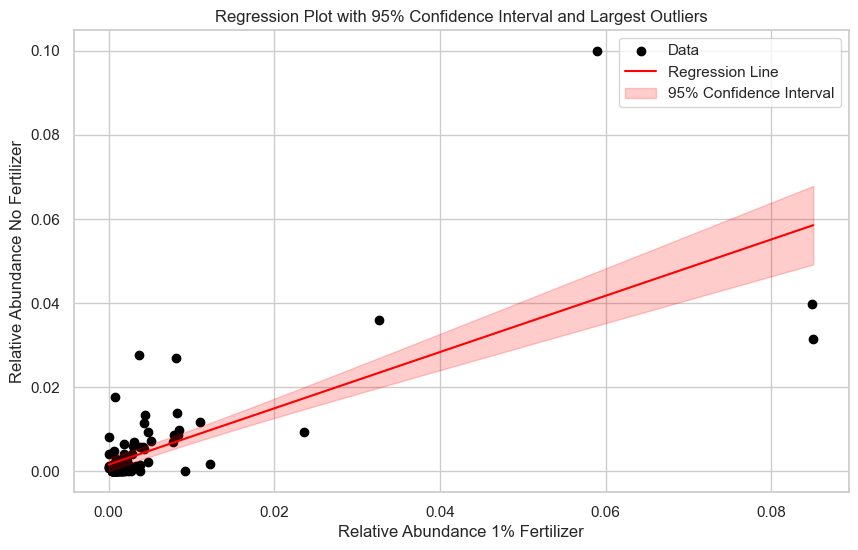
# Linear regression model
X = Combined['REL_ABUNDANCE_x']
y = Combined['REL_ABUNDANCE_y']
X_const = sm.add_constant(X) # Add constant for intercept
model = sm.OLS(y, X_const).fit()
# Predict values and confidence intervals
x_values = np.linspace(X.min(), X.max(), 100)
x_values_const = sm.add_constant(x_values)
y_pred = model.predict(x_values_const)
predictions = model.get_prediction(x_values_const)
confidence_interval = predictions.conf_int(alpha=0.05)
# Calculate residuals
Combined['residual'] = y - model.predict(X_const)
Combined['abs_residual'] = Combined['residual'].abs()
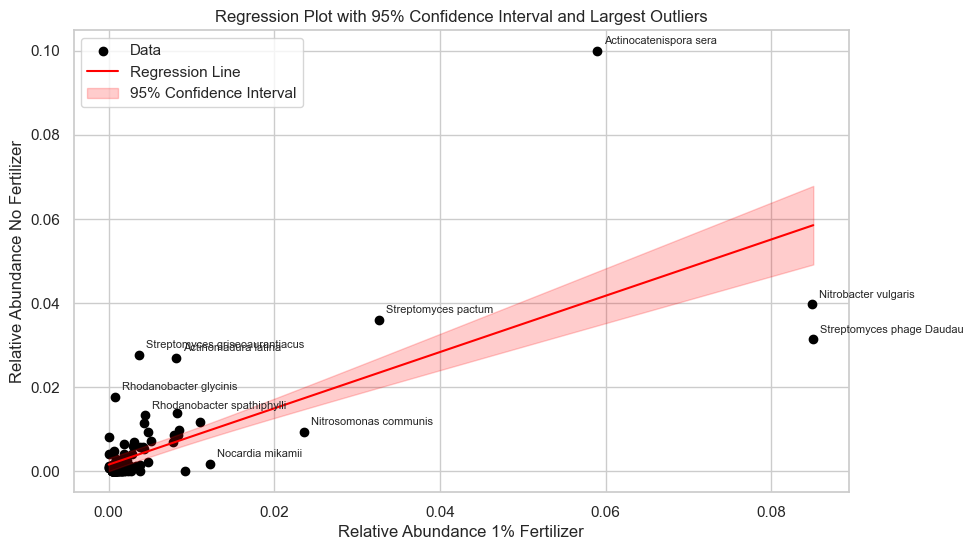
# Get the 10 largest outliers
largest_outliers = Combined.nlargest(10, 'abs_residual')
# Plot
sns.set(style="whitegrid")
fig, ax = plt.subplots(figsize=(10, 6))
# Scatter plot
ax.scatter(X, y, c='black', label="Data")
# Regression line
ax.plot(x_values, y_pred, color='red', label="Regression Line")
# Confidence interval
ax.fill_between(
x_values,
confidence_interval[:, 0], # Lower bound
confidence_interval[:, 1], # Upper bound
color='red',
alpha=0.2,
label="95% Confidence Interval"
)
# Customize plot
ax.set_xlabel("Relative Abundance 1% Fertilizer")
ax.set_ylabel("Relative Abundance No Fertilizer")
ax.set_title("Regression Plot with 95% Confidence Interval and Largest Outliers")
ax.legend()
plt.show()